![]() AJIT KUMAR PASSARI1, VINEET KUMAR MISHRA1, VIJAI KUMAR GUPTA2, RATUL SAIKIA3 and BHIM PRATAP SINGH1*
AJIT KUMAR PASSARI1, VINEET KUMAR MISHRA1, VIJAI KUMAR GUPTA2, RATUL SAIKIA3 and BHIM PRATAP SINGH1*
1Molecular Microbiology and Systematics Laboratory, Department of Biotechnology, Aizawl, Mizoram University, Mizoram, India
2Molecular Glyco-biotechnology Group, Department of Biochemistry, National University of Ireland Galway, Galway, Ireland
3Biotechnology Division, CSIR-North East Institute of Science and Technology, Jorhat, Assam, India
*Corresponding author: B.P. Singh, Department of Biotechnology, Aizawl, Mizoram University, Mizoram, India; e-mail: bhimpratap@gmail.com.
Submitted 28 March 2015, revised 22 June 2015, accepted 2 February 2016
DOI: 10.5604/17331331.1215611
Abstract
The prospective of endophytic microorganisms allied with medicinal plants is disproportionally large compared to those in other biomes. The use of antagonistic microorganisms to control devastating fungal pathogens is an attractive and eco-friendly substitute for chemical pesticides. Many species of actinomycetes, especially the genus Streptomyces, are well known as biocontrol agents. We investigated the culturable community composition and biological control ability of endophytic Streptomyces sp. associated with an ethanobotanical plant Schima wallichi. A total of 22 actinobacterial strains were isolated from different organs of selected medicinal plants and screened for their biocontrol ability against seven fungal phytopathogens. Seven isolates showed significant inhibition activity against most of the selected pathogens. Their identification based on 16S rRNA gene sequence analysis, strongly indicated that all strains belonged to the genus Streptomyces. An endophytic strain BPSAC70 isolated from root tissues showed highest percentage of inhibition (98.3 %) against Fusarium culmorum with significant activity against other tested fungal pathogens. Phylogenetic analysis based on 16S rRNA gene sequences revealed that all seven strains shared 100 % similarity with the genus Streptomyces. In addition, the isolates were subjected to the amplification of antimicrobial genes encoding polyketide synthase type I (PKS-I) and nonribosomal peptide synthetase (NRPS) and found to be present in most of the potent strains. Our results identified some potential endophytic Streptomyces species having antagonistic activity against multiple fungal phytopathogens that could be used as an effective biocontrol agent against pathogenic fungi.
Key words: Schima wallichii, biocontrol agent, endophytic Streptomyces, nonribosomal peptide synthetase (NRPS), polyketide synthase (PKS-I)
Introduction
Loss in crop production caused by fungal diseases is a major concern resulting in loss of 25% yield in western countries and almost 50% in developing countries (Gohel et al., 2006). Several economically important crops in tropical and sub-tropical regions are affected due to diverse fungal diseases, so it is important to control fungal diseases for constant food supply to an ever increasing world population (Oskey, 2009). Synthetic pesticides can keep fungal infections at an acceptable level. However, their extensive use can lead to several drawbacks such as environmental pollution, lack of specificity, development of resistant fungal strains or accumulation of compounds potentially hazardous to other life forms as well, including humans (Dahiya et al., 2006; Evangelista-Martinez, 2014). Therefore, control of fungal pathogens requires a more environmental friendly approach. Henceforth, microbe-based biocontrol methods could be an alternative to control devastating fungal diseases (Zhao et al., 2012; Patil et al., 2014). Microbial antagonists are commonly used for the biocontrol of fungal diseases by the use of various groups of microorganism like bacteria, algae, fungi and actinomycetes (Brimner and Boland, 2003).
Actinomycetes are characteristic as biological control agents against fungal plant pathogens by having the ability to produce secondary metabolites and biologically active compounds such as enzymes and antibiotics (Adegboye and Babalola, 2012; Mingma et al., 2014), of which genus Streptomyces is of meticulous importance (Evangelista-Martinez, 2014). Among the 10000 antimicrobial compounds produced by microorganisms, more than 50 % were isolated from actinomycetes and about 60% of the bioactive compounds developed for agricultural use originated from the genus Streptomyces (Anderson and Wellington, 2001). It is well known that Streptomyces sp. can produce a wide variety of secondary metabolites including antibiotics, enzymes and alkaloids, which may be the causative agent for antagonistic activity (Hayakawa et al., 1996).
Bioactive compounds such as benzoquinones (Rothrock and Gottlieb, 1984), aminoglycosides (Godfrey, 1995; Qin et al., 1994), polyenes (Smith et al., 1990; Raatikainen et al., 1994), nucleoside antibiotics (Hwang et al., 1994; Trejo-Estrada et al., 1998) are some agriculturally useful metabolites produced by genus Streptomyces. Several members of genus Streptomyces have been reported to significantly inhibit growth of plant fungal pathogens (Taechowisan et al., 2005; Maldonado et al., 2010; Evangelista-Martinez, 2014). To name some, Streptomyces rochei in combination with Trichoderma harzianum was used to control root rot in pepper (Ezziyyani et al., 2007), Streptomyces aureofaciens improved protection against anthracnose disease in mango caused by Colletotrichum goeosporioides (Haggag and Abdall, 2011) and Streptomyces griseus was used to control Fusarium wilt in tomato (Anitha and Rabeeth, 2009). Among the known commercial products produced by Streptomyces, Actinovate® and Mycostop® are the two most useful commercial products against foliar, root rot and wilt diseases marketed to date (Evangelista-Martinez, 2014).
In recent years, endophytic actinobacteria have attracted the attention of researchers as biocontrol agent against plant pathogens due to their better plant colonizing ability and antifungal activities. Their antagonistic capability has been proved against different phytopathogens, including Rhizoctonia solani, Verticillium dahlia, Fusarium oxysporum, Colletotrichum orbiculare (Coombs et al., 2004; Hasegawa et al., 2006; El-Tarabily et al., 2009; Shimizu et al., 2009).
Attempts were made to isolate actinomycetes as endophytes from various plants, where they live in symbiotic manner without causing any apparent damage to the host plant (Stone et al., 2000). Biological control ability of endophytic actinomycetes has been reported both in vitro and in vivo (Cao et al., 2005; Taechowisan et al., 2003). Evidences indicate that new endophytic actinomycetes were isolated from various organs of medicinal plants, and produced various bioactive compounds with a novel chemical structure, which would further increase the potential effectiveness as a biological control agent (Godfrey, 1995; Nimnoi et al., 2010). Therefore, there is a need for the isolation and characterization of actinomycetes from different geographical locations is important in order to identify new and commercially valuable genetic resources (Evangelista-Martinez, 2014). However, there is no study on the isolation of endophytic actinomycetes from medicinal plant Schima wallichii as biocontrol agents. Further, we used degenerate primers to amplify the antimicrobial genes like Polyketide synthase (PKS) type I and nonribosomal peptide synthetase (NRPS) because most of the biosynthetic pathways for the production of secondary metabolites are associates with these genes.
In our study, we have attempted to isolate endophytic actinomycetes from surface sterilized organs of medically important plant S. wallichii. The isolates were screened for their biocontrol ability against common fungal phytopathogens such as F. oxysporum, Fusarium proliferatum, Fusarium oxysporum f. sp. ciceri, Fusarium culmorum, Fusarium graminearum, Alternaria sp. and Colletotrichum sp. The potential strains selected as efficient biocontrol agents were identified by using 16S rRNA gene sequence. Further, biosynthetic potential of the potent strains was examined by amplification of PKS-I and NRPS genes.
Experimental
Materials and Methods
Sample collection. Healthy and disease-free tissues of the plant (S. wallichii) were collected from Dampa tiger reserve forest (23°44′N; 92°39′E) during the period of October 2013, based on ethanobotanical history, commonly used by the local tribes to heal the wounds caused by insects like spider and scorpion, as antiseptic and as external application in snake bite. Roots were dug out carefully so that sufficient amount of root material was collected. The tissues were placed in sterile bags and brought to the lab immediately and processed within 6 h of collection.
Surface sterilization and isolation of endophytic actinomycetes. Surface sterilization is the first and obligatory step for the recovery of true endophytes in order to kill the surface microbial population. The plant tissues were normally treated with oxidant or general sterilant for a period, followed by sterile rinse. The different plant tissues (leaf, fruit, root and bark) were used for isolation of endophytic actinomycetes and washed in running tap water for 5-10 minutes to remove adhered debris. The tissues were cut into small pieces (1.0 × 0.5cm) and surface sterilized by using three step procedures as described by Taechowisan and Lumyong (2003). Tissues were sterilized by sequential immersion in 0.1% Tween 20 for few seconds and transferred to clean conical flask, followed by 70% ethanol for 2 min, and a solution of 0.1% NaOCl for 1 min. Samples were washed in sterile water minimum three times to remove all surface disinfectants. An aliquot (0.1 ml) of the last wash was spreaded on starch casein agar (SCA) plates and incubated at 28 ±2°C for three weeks to proof the authenticity of the surface sterilization protocol (Kuster and Williams, 1964). The sterilized tissues were kept on autoclaved blotting paper to remove the any trace of water and inoculated on five different agar media viz. Starch Casein Nitrate Agar Media (SCNA), Actinomycetes Isolation Agar Media (AIA) and Tap Water Yeast Extract Agar Media (TWYE) at the rate of 10-15 tissue bits per plate. Nalidixic acid and Cycloheximide (80 ug/ml) were added to the media to inhibit the fungal and eubacterial growth. The plates were incubated at 28 ±2°C for 3-4 weeks, actinomycetes colonies were transferred and maintained on ISP2 media by repetitive streaking.
Identification of isolated endophytic actinomycetes. The isolates were identified based on cultural and morphological characteristics, including, colonies on the plate, aerial and substrate color, spore mass color, production of melanoid pigments and color of diffusible pigments (Goodfellow and Haynes, 1984; Simone et al., 1999). Based on Bergey’s manual of systematic bacteriology, we classified the isolates by looking onto the aerial and substrate mycelia color in the following series: gray, white, red, yellow, green, blue, and violet (Buchnan and Gibbons, 2000). Microscopic characteristics using light microscopy and gram-stain properties were also performed and observed that the spore chains under light microscope, showed various spore characters like straight or flexuous chains, spira, extended, long and open coils. The spore chain morphology and surface of spore were examined by field emission gun-scanning electron microscopy (FEG-SEM) of 10-day old cultures grown on ISP4 according to the method described previously Kumar et al. (2011). Different biochemical tests like starch hydrolysis, Citrate utilization test, Indole test, methyl red test, Vogus-Proskauer test, catalase test were performed to characterize actinomycetes till genus level.
In vitro antifungal bioassay. The endophytic actinomycetes isolates were evaluated for their antagonistic activity against seven major plant pathogenic fungi: F. oxysporum: CABI-293942; F. oxysporum f. sp. ciceri MTCC-2791, F. proliferatum: MTCC-286, F. culmorum MTCC-2090, F. graminearum MTCC-1893, Alternaria sp. MTCC-9601 and Colletotrichum sp. MTCC-3405 by dual-culture in vitro assay according to Bredholdt et al. (2007). All the pathogens were maintained on potato dextrose agar and maintained in a molecular microbiology and systematic laboratory, Department of Biotechnology, Mizoram University. An agar block of fungal pathogen was prepared by using sterile cork borer with diameter of 8 mm, and placed at the centre of PDA plate. Two endophytic actinomycetes discs (8 cm) 7 days old, grown on starch casein agar incubated at 28°C, were placed on the opposite sides of the plates, 3 cm away from the fungal block. Plates without endophytic actinomycetes discs were served as control. All plates were inoculated at 28°C for 14 days and percentage of inhibition was calculated by using the formula C-T/C x 100, where, C is the colony growth of fungal pathogen in control, and T is the colony growth in dual culture. All experiments were carried out in triplicates. Antagonistic activity was considered positive (+) if the zone of inhibition of the growth zone was more than 3 mm.
Molecular identification by using 16S rRNA gene amplification. The identity of the selected strains was determined based on the amplification and sequencing of 16S rRNA gene. Total genomic DNA was extracted by the Puregene Yeast/Bact Kit B (QIAGEN). The integrity of the genomic DNA was visualized by gel electrophoresis in 0.8% (w/v) by using the gel documentation system XR+ system (BioRed, Singapore).
Fragment of 16S rRNA gene were amplified by PCR using universal primers- PA: 5′-AGA GTT TGA TCC TGG CTC AG-3′) and PH: 5′-AAG GAG GTG ATC CAG CCG CA-3′ (Qin et al., 2009). The PCR reaction mixture was carried out in 25 µl total volume, containing 50 ng of genomic DNA, 2.5 µl (10 x) Taq Buffer, 1.5 µl (15 mM) MgCl2, 2.0 (2.5 mM) dNTPs, 0.5µl (10 pmol) each primer and 1 µl (1 U) Taq DNA Polymerase. The PCR conditions consisted of an initial denaturation at 94°C for 5 min, followed by 30 amplification cycles of denaturation at 94°C for 1 min, annealing at 57°C for 40 sec, extension at 72°C for 1 min and a final extension of 10 min at 72°C. The amplified PCR product was separated on 1.2 % agarose gel using TAE buffer and examined under gel documentation system XR+ (BioRed). The amplified amplicon was purified by using the Purlink PCR Purification Kit (In-vitrogen) and was sequenced commercially at SciGenom Labs Pvt. Ltd, India. Sequences were assembled and trimmed by using Finch TV version 1.4 (Geospiza inc.).
Phylogenetic analysis. Sequences of 16S rRNA gene were analyzed for homology using BlastN search program and very closely related species showing high level of identity (97%-100%) was considered as closest match. These sequences, along with other actinomycetes strains retrieved from NCBI GenBank (www.ncbi.nlm.nih.gov) were used for the construction of phylogenetic tree after pairwise aligned using ClustalW (Thompson et al., 1997). All the assembled sequences were submitted to NCBI Genebank and accession numbers were obtained. A Maximum-Likelihood Tree was constructed using Kimura 2-Parameter (Kimura, 1980) and reliability of phylogenetic tree was evaluated by bootstrap analysis with 1000 resamplings (Felsenstein, 1985) with Mega 5.05 software, taking Escherichia coli as outgroup.
PCR amplifications of antimicrobial genes (PKS-I and NRPS). Two antimicrobial genes Polyketide synthase (PKS) type I and nonribosomal peptide synthetase (NRPS) were amplified by using degenerate primers: K1F 5’-TSAAGTCSAACATCCGBCA-3’ and M6R 5’-CGCAGGTTSCSGTACCAGTA-3’ and A3F 5’-GCSTACSYSATSTACACSTCSGG-3’ and A7R 5’-SASGTCVCCSGTSGCGTAS-3’ respectively (Ayuso-Sacido and Genilloud, 2005). The PCR was performed in Veriti thermal cycler (Applied Biosystems, Singapore) in a final volume of 25 µl containing 25 ng of genomic DNA, 1.0 U of Taq DNA polymerase, 1 mM MgCl2, 0.5 mM of dNTPs, 1.0 µM of each primer and 10% DMSO. PCR conditions were one denaturation step at 95°C for 4 min., followed by 30 cycles of denaturation at 95°C for 60 s, annealing at 59°C for 60 s, and extension at 72°C for 2 min. Final extension step was done at 72°C for 10 min. A negative control reaction mixture without DNA template was also included with each set of PCR reactions. The PCR product was visualized on 1.5% agarose gel as stated above.
Statistical analysis. The data were calculated statically by using Microsoft Excel XP 2007 and significance difference (P ≤ 0.05) was estimated by one way analysis of variance (ANOVA) between antimicrobial activities of different isolates by using SPSS software version 16.0.
Results
The distribution and identification of endophytic actinomycetes associated with different organs of medicinal plant S. wallichii, collected from Dampa Tiger Reserve, the largest wildlife sanctuary in Mizoram, Northeast, India, along with their in vitro antifungal activities to test the ability of the isolates to suppress the growth of fungal pathogens was studied. Twenty two isolates from 68 tissues were preliminarily characterized morphologically and biochemically according to international Streptomyces project (ISP method) and by following Bergey’s manual of determinative bacteriology. The Scanning electron microscope (SEM) result showed that the aerial mycelia produce spiral spore chains (Fig. 1).

Isolated cultures were designated as BPSAC (2, 8, 16, 20, 32, 40, 42, 48, 54, 57, 60, 65-72, 75 and 81) (Table I and II). The distribution of endophytic actinomycetes was found to be highest in roots (n = 9, 40.9%) followed by bark (n = 6, 27.2%), leaves (n = 4, 18.1%) and fruit (n = 3, 13.6%). Five different nutritional media were used to understand the best suitable media for the recovery of endophytic actinomycetes and found that starch casein nitrate agar media (SCNA) (n = 8, 36.3%) was most effective followed by actinomycetes isolation agar media (AIA) (n = 6, 27.0%), tap water yeast extract agar media (TWYE) (n = 5, 22.0%), glycerol asparagine agar media (ISP5) (n = 2, 14.0%) and yeast and malt extract agar media (ISP2) (n = 1, 9.0%) (Fig. 2).
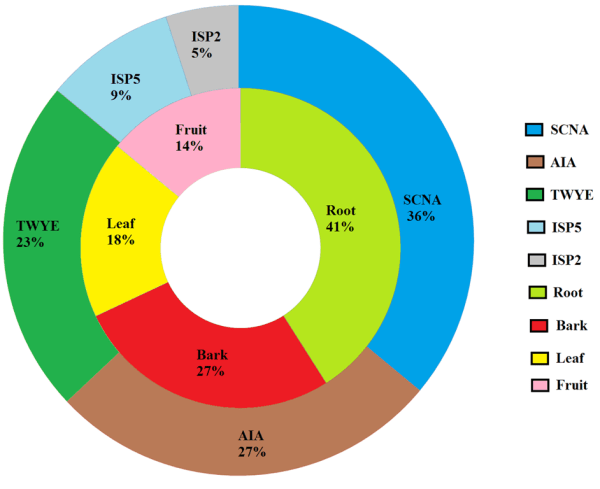
Table I
Morphological and microscopic characteristics of endophytic actinomycetes isolates with their different media
| Isolate No. and NCBI Genbank accession No. | Isolate identified | Growth and colony nature | Aerial Mycelia | Substrate Mycelia | Pigmentation | Media name |
| BPSAC2 | Streptomyces sp. | slow and rough | white | white | no | SCNA |
| BPSAC8 | Streptomyces sp. | slow and powdery | brownish white | light brown | no | ISP2 |
| BPSAC16 | Streptomyces sp. | slow and smooth | yellow | light yellow | no | ISP5 |
| BPSAC20 | Streptomyces sp. | slow and rough | orange | light orange | no | AIA |
| BPSAC22 | Streptomyces sp. | slow and powdery | brownish white | brown | yellowish brown | SCNA |
| BPSAC32 | Streptomyces sp. | slow and sticky | orange | light orange | no | TWYE |
| BPSAC40 | Streptomyces sp. | slow and rough | brownish white | brownish white | no | AIA |
| BPSAC42 | Streptomyces sp. | slow and firm | gray | light brown | no | SCNA |
| BPSAC48 | Streptomyces sp. | slow and sticky | orange | orange | no | SCNA |
| BPSAC54 | Streptomyces sp. | slow and rough | brownish white | brownish white | no | AIA |
| BPSAC57 | Streptomyces sp. | slow and powdery | white | white | no | SCNA |
| BPSAC60 | Streptomyces sp. | slow and smooth | yellow | light yellow | no | SCNA |
| BPSAC65 (KJ914903) |
S. sampsonii | slow and rough | brownish white | brownish white | yellowish brown | TWYE |
| BPSAC66 (KJ914904) |
S. olivaceus | slow and firm | brownish white | light brown | no | AIA |
| BPSAC67 | Streptomyces sp. | slow and powdery | light brown | brown | no | TWYE |
| BPSAC68 (KJ914906) | Streptomyces sp. | slow and sticky | yellow | yellow | no | TWYE |
| BPSAC69 (KJ914907) |
Streptomyces sp. | slow and rough | cream white | cream white | light brown | SCNA |
| BPSAC70 (KJ914908) |
Streptomyces sp. | slow and rough | brownish white | brown | light brown | AIA |
| BPSAC71 (KJ914909) |
S. tempisquensis | slow and powdery | gray | light brown | light brown | AIA |
| BPSAC72 (KJ914910) |
S. anulatus | slow and firm | light brown | light brown | no | TWYE |
| BPSAC75 | Streptomyces sp. | slow and sticky | yellow | yellow | no | SCNA |
| BPSAC81 | Streptomyces sp. | slow and rough | white | white | no | ISP5 |
Table II
Biochemical characterization of endophytic Actinomycetes isolates
| Isolate No | Indole | Catalase | Urease | Oxidase | Nitrate | Starch |
| BPSAC2 | + | + | + | + | + | + |
| BPSAC8 | – | + | + | – | + | – |
| BPSAC16 | – | – | + | + | – | – |
| BPSAC20 | – | – | + | – | + | + |
| BPSAC22 | – | + | – | + | – | + |
| BPSAC32 | – | + | – | + | – | – |
| BPSAC40 | – | + | + | + | + | – |
| BPSAC42 | – | + | + | – | – | – |
| BPSAC48 | – | – | + | + | + | + |
| BPSAC54 | – | – | – | – | + | – |
| BPSAC57 | – | – | + | – | + | – |
| BPSAC60 | – | – | – | + | – | – |
| BPSAC65 | + | + | + | – | + | + |
| BPSAC66 | – | + | + | + | + | + |
| BPSAC67 | – | + | – | – | + | – |
| BPSAC68 | – | + | + | + | – | – |
| BPSAC69 | – | + | – | + | – | – |
| BPSAC70 | + | + | + | + | + | + |
| BPSAC71 | + | + | + | + | + | + |
| BPSAC72 | + | + | + | + | – | + |
| BPSAC75 | – | + | + | – | + | + |
| BPSAC81 | + | + | + | + | – | – |
Fifteen of the 22 isolates showed significant growth inhibitory activity against at least two tested pathogens, when assayed against seven fungal phytopathogens (F. culmorum MTCC-2090, F. proliferatum MTCC-286, F. oxysporum CABI-293942, F. graminearum MTCC-1893, F. oxysporum f. sp. ciceri MTCC-2791, Alternaria sp. MTCC-9601 and Colletotrichum sp. MTCC- 3405). The results showed that most of the strains suppressed the test fungi with percentage of inhibition ranging from 26% to 98%. All isolates showed antagonistic activity against F. culmorum and F. proliferatum, whereas four isolates BPSAC (65, 68, 70 and 71) exhibited positive antagonistic effect against all selected pathogens (Fig. 3).

Antagonistic activity of isolate number BPSAC 70 showed most prominent antagonism against F. culmorum (98.3%) and F. graminearum (92.3%), whereas isolate numbers BPSAC 66, 68 and 71 showed the highest antagonistic activities against Alternaria sp. (90.6%), F. oxysporum f. sp. ciceri (92.3%) and F. proliferatum (90.6%), respectively. The use of several indicator organisms will help us in selecting the broad spectrum antifungal strains. Among all selected pathogens Alternaria sp. was the most susceptible fungal pathogen against most of the endophytic actinomycetes isolates with the exception to BPSAC (2, 22, 32, 42, 54, 60 and 69) (Table III).
Table III
Antifungal activity of endophytic actinomycetes against plant fungal pathogens
| Isolate No | Percentage of inhibition zone (PI) ± SD against | ||||||
| F. oxysporum CABI-293942 |
F. graminearum MTCC-1893 |
F. culmorum MTCC-2090 |
F. proliferatum MTCC-286 |
F. oxy. ciceri MTCC-2791 |
Alternaria sp. MTCC-9601 |
Colletotrichum sp. MTCC-3405 |
|
| BPSAC2 | 0.0±0.0a | 46.6±2.8a | 44.0±3.4a | 40.6±5.1a | 0.0±0.0a | 0.0±0.0a | 37.6±2.5a |
| BPSAC16 | 0.0±0.0a | 41.3±3.2bc | 37.6±2.5bc | 44.0±3.4bc | 0.0±0.0a | 35.3±3.2bc | 0.0±0.0bc |
| BPSAC22 | 40.6±5.1bc | 0.0±0.0bde | 35.3±3.2bde | 26.6±2.8bde | 31.3±3.2bc | 0.0±0.0a | 0.0±0.0bc |
| BPSAC32 | 0.0±0.0a | 0.0±0.0bde | 40.6±5.1bdfg | 37.6±2.5bdfg | 40.6±5.1bde | 0.0±0.0a | 37.1±2.8a |
| BPSAC40 | 0.0±0.0a | 0.0±0.0bde | 39.6±4.0bdfhi | 30.2±2.8bdfhi | 0.0±0.0a | 35.3±3.2bc | 0.0±0.0bc |
| BPSAC42 | 0.0±0.0a | 0.0±0.0bde | 36.3±3.2bdfhjk | 40.6±5.1a | 0.0±0.0a | 0.0±0.0a | 0.0±0.0bc |
| BPSAC54 | 0.0±0.0a | 0.0±0.0bde | 85.3±5.1bdfhjlm | 29.7±2.8bdfhi | 0.0±0.0a | 0.0±0.0a | 0.0±0.0bc |
| BPSAC60 | 44.0±3.4bde | 72.3±2.5bdfg | 44.0±3.4a | 30.7±2.8bdfhi | 31.3±3.2bc | 0.0±0.0a | 40.6±5.1bde |
| BPSAC65 | 90.0±2.0bdfg | 84.5±0.5bdfhi | 74.6±5.0bdfhjlno | 85.0±0.5bdfhjk | 75.3±2.5bdfg | 62.6±0.57bde | 73.3±0.57bdfg |
| BPSAC66 | 72.3±2.5bdfhi | 0.0±0.0bde | 64.6±0.5bdfhjlnpq | 79.0±1.0bdfhjlm | 72.3±2.5bdfhi | 90.6±0.5bdfg | 0.0±0.0bc |
| BPSAC68 | 85.3±5.1bdfhjk | 76.67±2.8bdfhjk | 72.3±2.5bdfhjlnprs | 72.3±2.5bdfhjlno | 92.3±2.5bdfhjk | 76.67±2.8bdfhi | 64.6±0.5bdfhi |
| BPSAC69 | 0.0±0.0a | 72.3±2.5bdfg | 72.3±2.5bdfhjlnprs | 76.67±2.8bdfhjlnpq | 79.0±1.0bdfhjlm | 0.0±0.0a | 52.5±0.5bdfhjk |
| BPSAC70 | 90.6±0.5bdfg | 92.3±5.1bdfhjlm | 98.3±2.8bdfhjlnprt | 86.0±1.7bdfhjlnprs | 76.67±2.8bdfhjln | 64.6±0.5bdfhjk | 74.6±5.0bdfhjlm |
| BPSAC71 | 74.6±5.0bdfhjlm | 85.3±2.5bdfhjlno | 74.6±5.0bdfhjlnotuv | 90.6±0.5bdfhjlnprtu | 44.0±3.4bdfhjlno | 56.6±2.8bdfhjlm | 72.3±2.5bdfhjlno |
| BPSAC72 | 72.3±2.5bdfhi | 85.3±5.1bdfhjlm | 76.67±2.8bdfhjlnprtuwx | 74.6±5.0bdfhjlnprtvw | 0.0±0.0a | 87.0±1.7bdfhjlno | 85.3±5.1bdfhjlnpq |
| Control | 35.4±2.5bdfhjln | 38.2±2.2bdfhjlnp | 30±0.0bdfhjlnprtuwy | 25.1±2.7bdfhjlnprtvx | 35.4±2.5bdfhjlnp | 28.0±2.3bdfhjlnp | 26.6±2.8bdfhjlnpr |
Mean (±SD) followed by the same letter(s) in each column are not significantly different at P < 0.05 using Duncan’s new multiple range test.
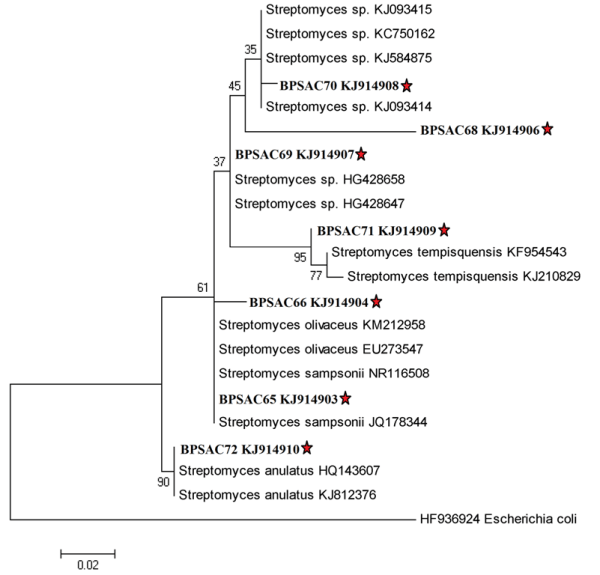
The selected potential isolates BPSAC (65, 66, 68, 69, 70, 71 and 72) which showed highest antagonistic activity against most of the pathogens were identified by amplification of 16S rRNA gene. The sequences were aligned by BLAST analysis along with the type strains downloaded from NCBI GenBank databases. Analysis of partial 16S rRNA gene sequences (565-830 bp) of potential seven strains exhibited high level of sequence similarity (97-99%) with sequences of Streptomyces species deposited in NCBI GeneBank. This indicates that all strains were closely related with the members of genus Streptomyces. The phylogenetic tree was constructed based on maximum likelihood method (Fig. 4) with Kimura 2-parameter model (R = 1.26) according to lowest BIC values using Mega 5.05. Gaps were treated by pairwise deletion and the estimated Transition/Transversion bias (R) was 1.25. The phylogenetic tree also confirmed the above results and the potent isolates were shorted out into four groups along with their closest relatives retrieved from NCBI GenBank. Isolates BPSAC (68, 69 and 70) had 96% – 99% similarity with type strains Streptomyces sp. and interestingly falls in one group. Other isolates conform to be a branch with Streptomyces sampsonii, Streptomyces olivaceus, Streptomyces tempisquensis and Streptomyces anulatus.
All twenty two isolates were subjected to the detection of PKS and NRPS genes and it was found that nine strains BPSAC (2, 32, 65, 66, 68, 69, 70, 71 and 72) were positive for the PKSI and 11 strains BPSAC (16, 32, 54, 60, 65, 66, 68, 69, 70, 71 and 72) were positive for NRPS gene fragments whereas other strains could not showed any amplification. Interestingly, all seven isolates which showed antagonistic activity, also showed the presence of PKS type I and NRPS gene fragments, which further proves them to be potential antagonistic strains and needs further attention (Fig. 5).

Discussion
Chemical-mediated suppression of plant pathogens is generally the primary method to repress the plant diseases. Indeed, the range of secondary metabolites produced by Streptomyces and other microorganism offers a great potential to fight many soil borne pathogens. As compared to Streptomyces in the rhizosphere, endophytic Streptomyces are expelled from the competition with other soil microorganisms and can efficiently colonize plant tissues. Thus, endophytic Streptomyces plays an important role in the development of plant by enhancing nutrient assimilation or by producing secondary metabolites (Kizuka et al., 2002). Endophytic Streptomyces have been investigated in many studies as biocontrol agent and revealed as a promising resource for agricultural industry (Cao et al., 2005; Shimizu et al., 2006). Previous studies have proved the use of endophytic Streptomyces as biocontrol agent against F. oxysporum (Sardi et al., 1992), bacteria, yeast and filamentous bacteria (Shimizu et al., 2001).
Plants growing in biodiversity rich areas with ethano-botanical history are likely to house endophytes with greater potential. Endophytes acquired a specific ability that allows them to sustain under the living tissues without any detectable infectious symptoms to the host. Thus, they are of immense importance to the host due to their capability to produce a wide array of natural bioactive compounds (Sardi et al., 1992; Strobel and Daisy, 2003; Cao et al., 2005; Kim et al., 2012).
However, this study was conducted for the first time to understand the endophytic actinomycetes population distribution and their potential as a biocontrol agent associated with traditional medicinal plant S. wallichii. In total twenty two isolates were obtained, among them maximum were isolated from root tissues (n = 9, 40.9%) followed by bark (n = 6, 27.2%), leaves (n = 4, 18.1%) and fruit (n = 3, 13.6%). Our results are in consensus with several studies, indicating the population of endophytic actinomycetes as highest in root tissues (De-Araujo et al., 2000; Taechowisan and Lumyong, 2003; Cao et al., 2005; Passari et al., 2015). To name a few, Verma et al. (2009) reported the highest percentage of endophytic actinomycetes was obtained from roots (55%) followed by other tissues of 20 different Azadirachta indica trees.
The reason may be due to the fact that actinomycetes present in rhizosphere can easily penetrate to root tissues and since plants collect nutrient and water through their roots, this may be the major source for the recovery of actinomycetes (Nimnoi et al., 2010). Though, Kayini and Pandey (2010) have reported the recovery of endophytic fungi from S. wallichii but, to the best of our knowledge, this is the first attempt made to understand the distribution of endophytic actinomycetes and their potential as biocontrol agent from S. wallichii. Five different nutrient media were used for the recovery of endophytic actinomycetes and found that maximum isolates (36.3%) used SCNA as a source of nutrition, which was in contradictory with the findings of Coombs and Franco (2003) and Khamna et al. (2009a; 2009b), who suggested that TWYE and HV agar medium were the best media for the isolation of endophytic actinomycetes.
All isolates were screened for their antagonistic ability against selected fungal phytopathogens and interestingly, seven isolates identified as Streptomyces sp. found to inhibit growth of most of the selected pathogens. Genus Streptomyces has an excellent track record for the discovery of bioactive metabolite and for the production of natural antibiotics (Baltz, 2006; Mingma et al., 2014).
Microbial metabolites may have an active role in resistance development by functioning as signals mediating cross-talk between the endophytes and their host (Graner et al., 2003). Since the endophytic actinomycetes were isolated from medicinal plant and that strains expected to produce a wide variety of antifungal and plant growth regulatory bioactive metabolites (Bredi, 1989; Franco and Cautinho, 1991), they can be exploited as novel sources of natural products and novel biocontrol agents as well. Endophytic actinomycetes are reported by many researchers for their role in plant protection against fungal pathogens. Two endophytic Streptomyces sp. were found to be active against all tested fungal pathogens including strains of Fusarium, Colletotrichum and Alternaria which was in agreement with earlier works of Khamna et al. (2009a; 2009b) and Intra et al. (2011). Streptomyces sp. were reported to suppress or inhibit plant pathogen F. oxysporum, the causative agent of cucumber Fusarium wilt (Zhao et al., 2012). Similarly, Verma et al. (2009) demonstrated the antifungal activity of endophytic actinomycetes against numbers of fungal pathogens.
Seven isolates were characterized by 16S rRNA gene sequence analysis and diversity of the potential isolates was found. The sequence of most isolates showed identity of 97% – 100% with BlastN sequences. All the isolates were classified as Streptomycetaceae family and identified as Streptomyces sp., S. sampsonii, S. tempisquensis, S. olivaceus and S. anulatus. Isolate BPSAC66 (S. olivaceus) was morphologically similar with BPSAC65 (S. sampsonii) and they were found to cluster together.
To understand the biosynthetic potential of the isolates, detection of genes encoding polyketide synthase and nonribosomal peptide synthetase mainly responsible for the synthesis of most biologically active polyketide and peptide compounds have been broadly used (Khamna et al., 2009a; 2009b). However, our results and findings from other researchers suggest that the antimicrobial potential of the culturable endophytic actinomycetes may only be assessed by screening of antimicrobial activity against desired pathogens. In our study, most of the isolates showed the presence of PKSI and NRPS genes, also showed antifungal activity against most of the tested pathogens which is contrary to the findings of Qin et al. (2009), who stated that antimicrobial activity results and the detection of functional genes showed no direct relationship.
Aknowledgement
This work was supported by grants from the DBT sponsored NER-Twinning project (No. BT/209/NE/TBP/2011), the Government of India, New Delhi. Authors are also thankful to the Department of Biotechnology, for establishment of DBT-BIF centre and DBT-state Biotech Hub in the Department, which has been used for the present study.
Literature
Adegboye M.F. and O.O. Babalola. 2012. Taxonomy and ecology of antibiotic producing actinomycetes. Afr. J. Agric. Res. 7:2255-2261.
Anderson A.S. and M.H.E. Wellington. 2001. The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol. 51:797-814.
Anitha A. and M. Rabeeth. 2009. Control of Fusarium wilt of tomato by bioformulation of Streptomyces griseus in green house condition. Afr. J. Basic Appl. Sci. 1:9-14.
Ayuso-Sacido A. and O. Genilloud. 2005. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 49:10-24.
Baltz R.H. 2006. Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 33:507-513.
Berdi J. 1989. The discovery of new bioactive metabolites; screening and identification. Prog. Ind. Microbiol. 27:3-25.
Bredholdt H., O.A. Galatenko, K. Engelhardt, E. Fjaervik, L.P. Terekhova and S.B. Zotchev. 2007. Rare actinomycete bacteria from the shallow water sediments of the Trondheim fjord, Norway: isolation, diversity and biological activity. Environ. Microbiol. 9:2756-2764.
Brimner T.A. and G.J. Boland. 2003. A review of the non-target effects of fungi used to biologically control plant diseases. Agr. Ecosyst. Environ. 100:3-16.
Buchnan R.E. and N.E. Gibbons. 2000. In: Bergey’s manual of determinative bacteriology. 9th ed. Actinomycetales.
Cao L., Z. Qiu, J. You, H. Tan and S. Zhou. 2005. Isolation and Characterization of endophytic Streptomycete antagonists of Fusarium wilt pathogen from surface-sterilized banana roots. FEMS Microbiol. Lett. 247:147-52.
Coombs J.T. and C.M.M. Franco. 2003. Isolation and identification of actinobacteria from surface sterilized wheat roots. Appl. Env. Microbiol. 69:5603-5608.
Coombs J.T., P.P. Michelsen and C.M.M. Franco. 2004. Evaluation of endophytic actinobacteria as antagonists of Gaeumannomyces graminis var. tritici in wheat. Biol. Control. 29:359-366.
Dahiya N., R. Tewari and G.S. Hoondal. 2006. Biotechnological aspects of chitinolytic enzymes. Appl. Microbiol. Biotechnol. 71:773-782.
De-Araujo M.J., C.A. Silva and J.L. Azevedo. 2000. Isolation of endophytic actinomycetes from roots and leaves of Maize (Zea mays). Braz. Arch. Biol. Techn. 43:447-51.
El-Tarabily K.A., A.H. Nassar, G.E. Hardy and K. Sivasithamparam. 2009. Plant growth promotion and biological control of Pythium aphanidermatum, a pathogen of cucumber, by endophytic actinomycetes. J. Appl. Microbiol. 106:13-26.
Evangelista-Martinez Z. 2014. Isolation and characterization of soil Streptomyces species as potential biological control agents against fungal plant Pathogens. World J. Microbiol. Biotechnol. 30:1639-1647.
Ezziyyani M., M.E. Requena, C. Egea-Gilabert and M.E. Candela. 2007. Biological control of Phytophthora root rot of pepper using Trichoderma harzianum and Streptomyces rochei in combination. J. Phytopathol. 155:342-349.
Felsenstein J. 1985. Confidence limits of phylogenies: An approach using the bootstrap. Evol. 39:783-791.
Franco C.M. and L.E. Cautinho. 1991. Detection of novel secondary metabolites. Crit. Rev. Biotechnol. 11:193-276.
Godfrey C.R.A. 1995. Agrochemicals from Natural Products. New York, Marcel Dekker.
Gohel V., A. Singh, M. Vimal, P. Ashwini and H.S. Chhatpar. 2006. Bioprospecting and antifungal potential of chitinolytic Microorganisms. Afr. J. Biotechnol. 5:54-72.
Goodfellow M. and J.A. Haynes. 1984. Actinomycetes in marine sediments, pp. 453-472. In: Ortiz-Ortiz L., L.F. Bojalil and V. Yakoleff (eds). Biological, biochemical and biomedical aspects of actinomycetes. Academic Press, New York.
Graner G., P. Persson, J. Meijer and S. Alstrom. 2003. A study on microbial diversity in different cultivars of Brassica napus in relation to its wilt pathogen, Verticillium longisporum. FEMS Microbiol. Lett. 224:269-276.
Haggag W.M. and A.M. Abdall. 2011. Foliar application of Streptomyces aureofaciens improve protection in Mango against post-harvest anthracnose and enhances fruit yield. Eur. J. Sci. Res. 63:139-149.
Hasegawa S., A. Meguro, M. Shimizu, T. Nishimura and H. Kunoh. 2006. Endophytic actinomycetes and their interactions with host plants. Actinomycetologica 20:72-81.
Hayakawa M., Y. Momose, T. Yamazaki and H. Nonomura. 1996. A method for the selective isolation of Microtetraspora glauca and related fourspored actinomycetes from soil. J. Appl. Bacteriol. 80:375-386.
Hwang B.C., S.J. Ahn and S.S. Moon. 1994. Production, purification and antifungal activity of the antibiotic nucleoside, tubercidin, produced by Streptomyces violaceusniger. Can. J. Bot. 72:480-485.
Intra B., I. Mungsuntisuk, T. Nihira, Y. Igarashi and W. Panbangred. 2011. Identification of actinomycetes from plant rhizospheric soils with inhibitory activity against Colletotrichum sp. the causative agent of anthracnose disease. BMC Res. Notes 4:98.
Kayini A. and R.R. Pandey. 2010. Phyllosphere Fungi of Alnus nepalensis, Castanopsis hystrix and Schima walichii in a Subtropical Forest of North East India. J. Am. Sci. 6:118-124.
Khamna S., A. Yokota and S. Lumyong. 2009a. Actinomycetes isolated from medicinal plant rhizosphere soil: diversity and screening of antifungal compound, indole-3-acetic acid and siderophore production. World J. Microbiol. Biotechnol. 25:649-655.
Khamna S., A. Yokota, J.F. Peberdy and S. Lumyong. 2009b. Antifungal activity of Streptomyces sp. isolated from rhizosphere of Thai medicinal plants. Int. J. Integr. Biol. 6:143-147.
Kim T.U., S.H. Cho, J.H. Han, Y.M. Shin, H.B. Lee and S.B. Kim. 2012. Diversity and physiological properties of root endophytic actinobacteria in native herbaceous plants of Korea. J. Microbiol. 50:50-57.
Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120.
Kizuka M., R. Enokita, K. Takahashi, Y. Okamoto, T. Otsuka, Y. Shigematsu, Y. Inoue and T. Okazaki. 2002. Studies on actinomycetes isolated from plant leaves: New plant growth inhibitors A-79197-2 and -3 from Dacthylosporangium aurantiacum SANK 61299. Actinomycetologica 16:14-16.
Kumar V., A. Bharti, O. Gusain and G.S. Bisht. 2011. Scanning electron microscopy of Streptomyces without use of any chemical fixatives. Scanning 33:1-4.
Kuster E. and S. Williams. 1964. Selection of media for isolation of Streptomycetes. Nat. 202:928-929.
Maldonado M.C., C.E. Orosco, M.A. Gordillo and A.R. Navarro. 2010. In vivo and in vitro antagonism of Streptomyces sp. RO3 against Penicillium digitatum and Geotrichum candidum. Afr. J. Microbiol. Res. 4:2451-2556.
Mingma R., W. Pathom-aree, S. Trakulnaleamsai, A. Thamchaipenet and K. Duangmal. 2014. Isolation of rhizospheric and roots endophytic actinomycetes from Leguminosae plant and their activities to inhibit soybean pathogen, Xanthomonas campestris pv. Glycine. World J. Microbiol. Biotechnol. 30:271-280.
Nimnoi P., N. Pongsilp and S. Lumyong. 2010. Endophytic actinomycetes isolated from Aquilaria crassna Pierre ex Lec and screening of plant growth promoters production. World J. Microbiol. Biotechnol. 26:193-203.
Oskay M. 2009. Antifungal and antibacterial compounds from Streptomyces strains. Afr. J. Biotechnol. 8:3007-3017.
Passari A.K., Mishra V.K., Saikia R., Gupta V.K. and Singh B.P. 2015. Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 6:273.
Patil N.N., M.S. Waghmode, P.S. Gaikwad, M.H. Gajbhiye, A.B. Gunjal, N.N. Nawani and B.P. Kapadnis. 2014. Potential of Microbispora sp. V2 as biocontrol agent against Sclerotium rolfsii the causative agent of southern blight of Zea mays L (Baby corn) in vitro studies. Indian J. Exp. Biol. 52:1147-1151.
Qin S., J. Li, H.H. Chen, G.Z. Zhao and W.Y. Zhu. 2009. Isolation, diversity and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microbiol. 75:6176-6186.
Qin Z., V. Peng, X. Zhou, R. Liang, Q. Zhou, H. Chen, D.A. Hopwood, T. Keiser and Z. Deng. 1994. Development of a gene cloning system for Streptomyces hygroscopicus var yingchengensis, a producer of three useful antifungal compounds by elimination of three barriers to DNA transfer. J. Bacteriol. 176:2090-2095.
Raatikainen O.J., T.H. Paivinen and R.T. Tahvonen. 1994. HPLC separation and subsequent detection of aromatic heptaene polyenes in peat after treatment with Streptomyces griseoviridis. Pestic. Sci. 41:149-154.
Rothrock C.S. and D. Gottlieb. 1984. Role of antibiosis in antagonism of Streptomyces hygroscopicus var geldanus to Rhizoctonia solani in soil. Can. J. Microbiol. 30:1440-1447.
Sardi P., M. Saracchi, S. Quaroni, B. Petrolini, G.E. Borgonovi and S. Merli. 1992. Isolation of endophytic Streptomyces strains from surface-sterilized roots. Appl. Environ. Microbiol. 58:2691-2693.
Shimizu M., T. Furumai, Y. Igarashi, H. Onaka, T. Nishimura, R. Yoshida and H. Kunoh. 2001. Association of induced disease resistance of rhododendron seedlings with inoculation of Streptomyces sp. R-5 and treatment with actinomycin D and amphotericin B to the tissue-culture medium. J. Anti. Biot. 54:501-505.
Shimizu M., A. Meguro, S. Hasegawa, T. Nishimura and H. Kunoh H. 2006. Disease resistance induced by nonantagonistic endophytic Streptomyces sp. on tissue-cultured seedlings of rhododendron. J. Gen. Plant Pathol. 72:351-354.
Shimizu M., S. Yazawa, Y. Ushijima. 2009. A promising strain of endophytic Streptomyces sp. for biological control of cucumber anthracnose. J. Gen. Plant Pathol. 75:27-36.
Simone D.G.P., M.M. Rosana, R.R.C. Rosalie, N.L.M. Maria, H.B. Marta and B.V. Alane. 1999. Influence of growth medium in protease and pigment production in Streptomyces cyzneus. Mem. Inst. Oswwaldo. Cruz. 94:173-177.
Smith J., A. Putnam and M. Nair. 1990. In vitro control of Fusarium diseases of Asparagus officinalis L. with a Streptomyces or its polyene antibiotic, Faeriefungin. J. Agric. Food Chem. 38:1729-1733.
Stone J.K., C.W. Bacon and J.F. White. 2000. An overview of endophytic microbes: endophytism defined, pp. 3-29. In: Bacon C.W. and J.F. White (eds). Microbial endophytes. Marcel Dekker, Inc., New York.
Strobel G.A. and B. Daisy. 2003. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67:491-502.
Taechowisan T., C. Lu, Y. Shen and S. Lumyong. 2005. Secondary metabolites from endophytic Streptomyces aureofaciens CMUAc130 and their antifungal activity. Microbiology 151:1691-1695.
Taechowisan T. and S. Lumyong. 2003. Activity of endophytic actinomycetes from roots of Zingiber officinale and Alpinia galanga against phytopathogenic fungi. Ann. Microbiol. 53:291-98.
Taechowisan T., J.F. Peberdy and S. Lumyong. 2003. Isolation of endophytic actinomycetes from selected plants and their antifungal activity. World J. Microbiol. Biotechnol. 19:381-385.
Thompson J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin and D.G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analys is tools. Nucleic Acids Res. 24:4876-82.
Trejo-Estrada S.L., A. Paszczynski and D.L. Crawford. 1998. Antibiotics and enzymes produced by biological agent Streptomyces violaceusniger YCED-9. J. Ind. Microbiol. Biotechnol. 21:81-90.
Verma V.C., S.K. Gond, A. Kumar, A. Mishra, R.N. Kharwar and A.C. Gange. 2009. Endophytic actinomycetes from Azadirachta indica A. Juss.: isolation, diversity, and anti-microbial activity. Microb. Ecol. 57:749-756.
Zhao S., C.M. Du and C.Y. Tian. 2012. Suppression of Fusarium oxysporum and induced resistance of plants involved in the biocontrol of cucumber Fusarium wilt by Streptomyces bikiniensis HD-087. World J. Microbiol. Biotechnol. 28:2919-2927.
